Title: Synchrotron X-Ray Diffraction and Pair Distribution Function Analysis of Drug/Polymer Dispersions: A Comparison of Subtraction Techniques to Isolate Intra- and Intermolecular Interactions
Authors: PA Smitha and SR Byrna, GLB de Araujob, CJ Benmorec
Affiliations: a) Improved Pharma LLC, b) Department of Pharmacy, Faculty of Pharmaceutical Sciences, University of Sao Paulo, c) X-ray Science Division, Advanced Photon Source, Argonne National Laboratory
Purpose: The pair distribution function (PDF) of an amorphous dispersion represents the sum of all atom-atom contacts in that dispersion. Difference pair distribution functions are obtained by subtracting the PDFs of the intramolecular drug and polymer contributions from the overall dispersion PDF. This approach provides important information on the structure of the dispersion and the presence or absence of domains of drug in that dispersion. The overall aim of this study is to compare results from a new subtraction approach to results obtained in published studies. Specifically, intramolecular and intermolecular reference PDF curves created with xINTERPDF1,2 from amorphous G(r) experimental data will be compared to those fit to S(Q) experimental data using XISF3 and methodology described previously.4 The ultimate goal is to compare these two approaches and illustrate their consistency.
Methods: Data collected from various spray-dried dispersions (1:3, 1:1, 3:1) of lapatinib/polymer5 or flubendazole/polymer6 were used in this study. Intramolecular and intermolecular PDF curves and the subtraction results obtained in those publications via the S(Q) files were used as-is. Conversely, the G(r) data created for those publications was used with xINTERPDF to obtain intramolecular and intermolecular PDF curves. The created intramolecular API PDF patterns were subtracted from the experimental G(r) data files, as were the polymer PDF curves. The subtraction results were then compared to the previous results. Secondly, to investigate the impact of data input for the molecular structure file, indomethacin was used as a test-case. Both alpha and gamma crystal structures were used in xINTERPDF, and the resulting calculated intramolecular and intermolecular indomethacin files were compared.
Results:
The xINTERPDF subtraction results on the lapatinib/polymer dispersions are displayed in Figures 1 and 2. All samples except the 1:3 lapatinib/HPMCP samples have evidence of API domains, as indicated by the broad peak ~4.3Å. These results are equivalent to those obtained previously. The exercise repeated for the flubendazole/polymer dispersions also confirmed the previously obtained results. Interestingly, the study involving different crystalline structure inputs for indomethacin resulted in significantly different calculated intramolecular and intermolecular PDFs. The calculated intramolecular PDFs are displayed in Figure 3, along with the experimental PDF of amorphous indomethacin.
Conclusions:
xINTERPDF is a viable and acceptable program for creating intramolecular and intermolecular PDF patterns. The program was successfully used on a variety of previously analyzed spray-dried drug/polymer dispersions, and subtractions were performed yielding equivalent results as before. Evidence of intramolecular API interactions indicating the presence of API domains were found in many of the dispersions, which led to a decrease in stability and subsequent crystallization. In each case study, the same drug/polymer dispersion was found to contain no API domains, no matter which data analysis approach was used. Secondly, the impact of reference data used as inputs into the program were examined for indomethacin with respect to selecting the proper molecular structure for PDF calculations. The results obtained from using the crystal structure file for alpha versus gamma indomethacin yielded significantly different results. Therefore, this type of PDF analysis could be very helpful in determining which conformation or mixture of conformations are present in the amorphous phase.
References:
- Shi, C, xINTERPDF, a graphical user interface for analyzing intermolecular pair distribution functions of organic compounds from X-ray total scattering data. J. Appl. Crystallogr, 51, 1498-1499 (2018).
- Juhas, P, Farrow, C. L., Yang, X., Knox, K. R., Billinge, S. J. L. Complex modeling: a strategy and software program for combining multiple information sources to solve ill posed structure and nanostructure inverse problems, Acta Crystallogr A. 71, 562-568 (2015).
- Mou, Q., Benmore, C. J., Yarger, J. L. X-ray Intermolecular Structure Factor (XISF): separation of intra-and intermolecular interactions from total X-ray scattering data. J. Appl. Crystallogr. 48, 950-952 (2015).
- Benmore, C. J. et al. Structural Characterization and Aging of Glassy Pharmaceuticals made Using Acoustic Levitation. J. Pharm. Sci. 102, 1290–1300 (2013).
- de Araujo, G. L. B., Benmore, C. J. & Byrn, S. R. Local Structure of Ion Pair Interaction in Lapatinib Amorphous Dispersions characterized by Synchrotron X-Ray diffraction and Pair Distribution Function Analysis. Sci. Rep. 7, 46367 (2017).
- de Araujo, G. L. B., Hydroxypropyl Methylcellulose Derivatives Amorphous Flubendazole Dispersions: Formulation Stability Assisted Through Pair Distribution Function Analysis, in press (2019).
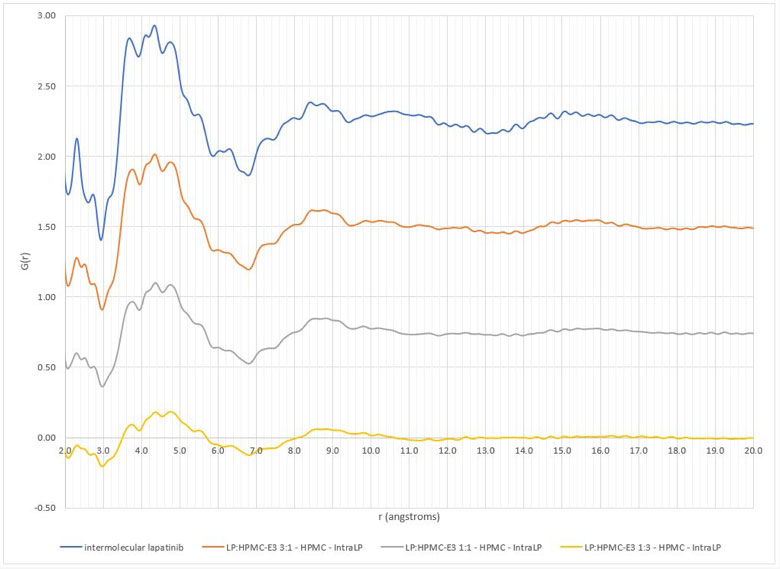
Figure 1: Comparison of intermolecular differential pair distribution functions of lapatinib/HPMC-E3 dispersions (top to bottom: pure lapatinib, 3:1 lapatinib:HPMC-E3, 1:1 lapatinib:HPMC-E3, 1:3 lapatinib/HPMC-E3)
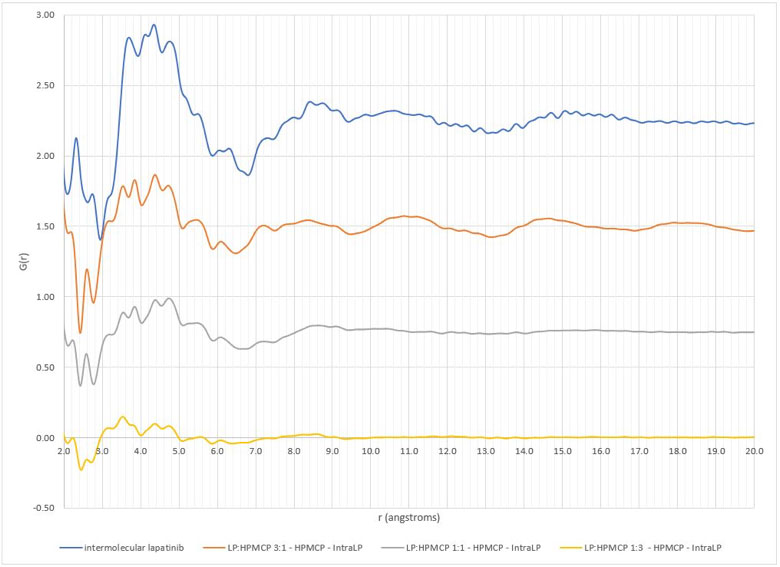
Figure 2: Comparison of intermolecular differential pair distribution functions of lapatinib/HPMCP dispersions (top to bottom: pure lapatinib, 3:1 lapatinib:HPMCP, 1:1 lapatinib:HPMCP, 1:3 lapatinib/HPMCP)
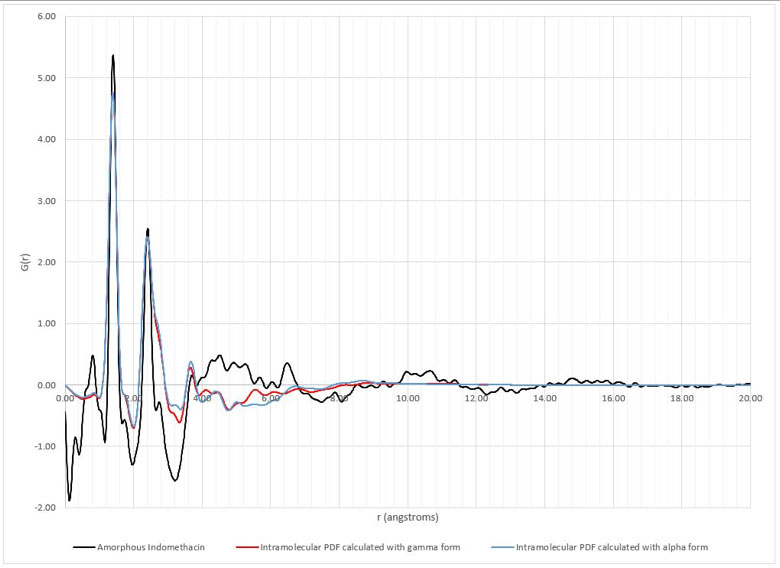
Figure 3: Intramolecular pair distribution functions of indomethacin created in xINTERPDF using either the gamma or the alpha form compared to the experimental pair distribution function of amorphous indomethacin
